More Information
Submitted: September 06, 2022 | Approved: September 15, 2022 | Published: September 16, 2022
How to cite this article: Yaqoob MD, Khawaja MAA, Amjad Q, Waseem A, Kanwal K, et al. Diagnostic evaluation of nasopharyngeal swab and saliva kits against SARS-CoV-2: Adequate rapid screening is deemed necessary to overcome COVID-19 Pandemic. Arch Biotechnol Biomed. 2022; 6: 010-013.
DOI: 10.29328/journal.abb.1001032
Copyright License: © 2022 Yaqoob MD, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: SARS-CoV-2; COVID-19; PCR; RDT kits; Nasopharyngeal swab; Saliva-based kits
Diagnostic evaluation of nasopharyngeal swab and saliva kits against SARS-CoV-2: Adequate rapid screening is deemed necessary to overcome COVID-19 Pandemic
Muhammad Danish Yaqoob1,2, Muhammad Abdul Ahad Khawaja1,3, Qurat-ul-Ain Amjad1,4, Atika Waseem1,4, Kashmala Kanwal1,4, Haleema Nadeem1,4, Madiha Munir1,4, Syeda Mushiat Zahra1,4, Zahra Zahid Piracha1 and Umar Saeed1*
1International Center of Medical Sciences Research (ICMSR), Islamabad (44000), Pakistan
2Binzhou Medical University, Yantai, Shandong, PR China
3King Edward Medical University, Lahore, Pakistan
4International Islamic University, Islamabad, Pakistan
#All authors contributed equally
*Address for Correspondence: Dr. Umar Saeed, International Center of Medical Sciences Research (ICMSR), Islamabad (44000), Pakistan, Email: [email protected], [email protected]
SARS-CoV-2 is the virus associated with the disease called COVID-19 and become a global pandemic. The only way to prevent its severe scenarios is through timely and rapid testing. In comparison to more time taking gold-standard RT-PCR testing, rapid diagnostic kits are used. For better prevention and diagnosis of SARS-CoV-2, the analysis of rapid diagnostic kits' accuracy and specificity is necessary. This study is meant to assess and examine the viability, responsiveness, and explicitness of quick antigen distinguishing nasopharyngeal swabs (NPS), and saliva-based units. The study was conducted on 200 suspected COVID-19 patients from Islamabad, 100 of which were RT-PCR positive while 100 were RT-PCR negative. For the analysis of Rapid diagnostic COVID-19 kits (RDT), nasopharyngeal swabs (NPS) and saliva samples were taken from the RT-PCR positive and negative patients. Among 100 RT-PCR positive patients, 62% were males (19 - 91 years), 34% were females (20 - 78 years) and 4% were children (6 - 17 years). False-negative results were significantly more observed in saliva-based RDTs of the sample (49%) as compared to nasopharyngeal swab RDT (38%). There were 2% invalid results in saliva-based RDT and 3% invalid results in Nasopharyngeal swab RDT. While among 100 RT-PCR negative patients 69% were males (19 - 80 yrs), 27% were females (18 – 77 yrs) and 4% were children (12 - 16 yrs.). False positive results were significantly more in saliva-based RDT (22%) as compared to Nasopharyngeal swab RDT (13%). The sensitivity and specificity of saliva-based RDT were 67% and 87% respectively while that of Nasopharyngeal swab (NPS) was 72% and 82% respectively, both of which were less than the gold standard RT-PCR sensitivity demanding the introduction of more sensitive RDT kits in Pakistan for accurate detection of COVID-19.
The seventh human coronavirus is known as a severe acute respiratory syndrome (SARA-CoV-2) first outbreak in Wuhan, Hubie province, China [1,2]. The deadly virus spread rapidly all over the world and infected 4,806,299 people and caused 318,599 deaths as of 20 May 2020 [3]. The deadly SARS-CoV-2 virus is a beta coronavirus and its subgenus is Sarbecovirus [4]. There were 43,820,929 positive cases of SARS-CoV-2 globally and were rising rapidly and the number of deaths in that period was 1,165,189, India and United States were the most badly influenced by COVID-19 [5]. On July 22, 2020, there were more than 1,47,65,256 confirmed cases of COVID-19 infection, and there were more than 6,12,054 fatalities in 200 countries (mortality rate of about 3.7%) [6].
Genomes investigation and examination with recently known COVID genomes show that SARS-CoV-2 presents exceptional highlights that discern it from other COVIDs: ideal partiality for angiotensin changing over catalyst 2 (ACE2) receptor and a polybasic cleavage site at the S1/S2 spike intersection that decides infectivity and host range [7,8]. The novel SARS-CoV-2, on the other hand, has an RNA genome size of 29.9 kb [9]. SARS-CoV-2 exhibits 88 % nucleotide sequence identity to the two SARS-like coronaviruses generated from bats (bat-SL-CoVZC45 and bat-SL-CoVZXC2), as well as 79% and 50% similarity to the SARS-CoV and MERS-CoV, respectively [10]. A rising number of publications suggest that during the process of geographical diffusion, the SARS-CoV2 genome has undergone evolutionary alterations and heterogeneity. Global SARS-CoV-2 isolates' pan-genomic study has identified numerous genomic areas with higher genetic variation and a distinctive mutation pattern [11].
Since a large number of infected cases were asymptomatic [12]. For the early diagnosis of cases, controlling the pandemic situation large-scale testing was proposed to be crucial. The availability of the complete genome of the SARS-CoV-2 virus within outbreak declaration two weeks, allowed the production and introduction of a broad range of RT-PCR kits by numerous developers and manufacturers. The process of Emergency Use Authorization (EUA) was used for the clinical application of these kits by regulatory agencies to deal with the demand of large-scale testing, instead of using the basic authorization process of granting full clearance for diagnostic applications [13,14].
To avoid viral pathogenicity early detection and isolation of affected cases were necessary [15]. The globally recommended and approved gold standard for SARS-CoV-2 detection and analysis refers to the nasopharyngeal swab (NPS) which is followed by real-time reverse-transcription polymerase chain reaction (RT-PCR) of the RNA extracted from the suspect [16,17]. But in the pandemic situation analyzing large-scale cases using RT-PCR within a small time limit was challenging and burdensome demanding for the other fast, correct and cost-efficient diagnosis of SARS-CoV-2 for resource-limited countries like Pakistan to fulfill national and international requirements [18]. Although there may be some doubt about the validity and efficacy of these assays in the real world, Rapid diagnostic kits (RDTs) for COVID-19 are cost-efficient, easy, and safe to use [19]. The NPS method is intrusive, potentially bleeding, and there is an increased likelihood that SARS-CoV-2 may be transmitted to healthcare employees [20]. While the collecting of saliva samples is safe to handle outside of hospitals and is non-intrusive [21]. Additionally, taking saliva samples on one's own can lower the risk of healthcare workers being transmitted with SARS-CoV-2 than NPS [22]. Notably, there was no discernible difference in the viral level of SARS-CoV-2 in NPS or saliva samples [23].
The current study was conducted by the International Center of Medical Sciences Research (ICMSR), Islamabad (44000), Pakistan, after getting IRB approval, from 1st Jan 2022 to 30th April 2022, as per standard operating procedures described previously [24]. Finding the best reliable diagnostic assay for SARS-CoV-2 RDTs based on saliva (INVBIO, INV-1047) or NPS (INVBIO, INVBIO-COVID) is difficult without sacrificing the validity of test results. We aimed to evaluate the specificity and sensitivity of NPS and Saliva based kits used in Pakistan for COVID-19 diagnosis, which could be helpful to formulate effective testing procedures for SARS-CoV-2 in Pakistan.
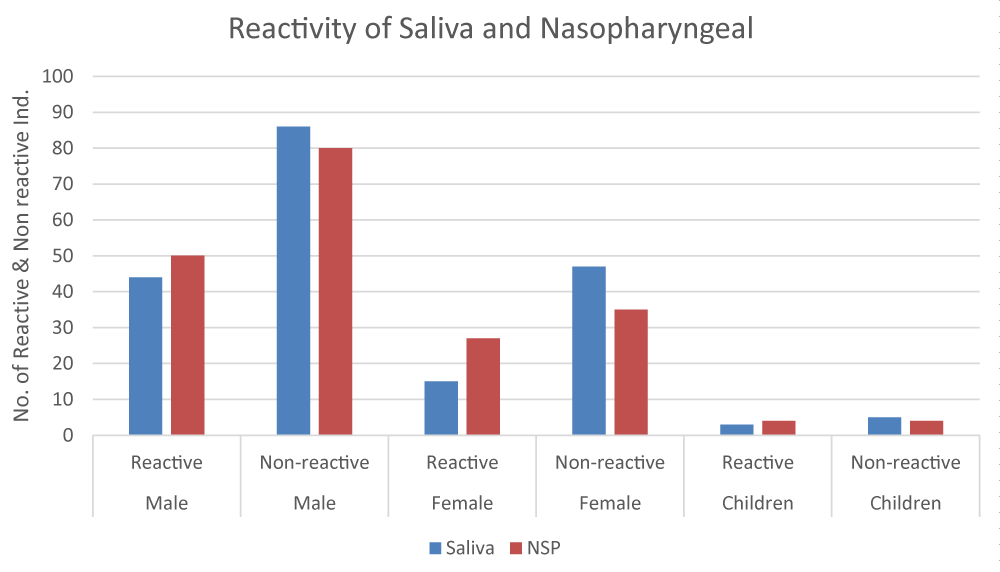
A total of 200 COVID-19 suspected patients were selected for evaluation of SARS-CoV-2 to antigen rapid test kits. Among selected individuals 130 (65%) were males, 62 (31%) were females and 8 (4%) were children. The average age of individuals was 41.5 years. Out of 200 patients, 99 showed positive results on saliva-based RT PCR i.e mean CT value less than 40, but on saliva-based antigen rapid test kits 62 (44 males, 15 females, and 3 children) individuals showed reactivity i.e positive results. In comparison with RT PCR for saliva, antigen rapid test kits for saliva showed sensitivity and specificity of 67% and 87% respectively, as shown in Table 1 and Figure 1.
| Table 1: Characteristics of patients and clinical features. | |||||
| Samples | Age (av) | RT-PCR Saliva | RT-PCR NSP | Saliva-ARTK | NSP-ARTK |
| Male (n =130) | 42.84 (19-91) | Reactive (n =62) Non-reactive (n =68) |
Reactive (n =62) Non-reactive (n =68) |
Reactive (n =44) Non-reactive (n =86) |
Reactive (n =50) Non-reactive (n =80) |
| Female (n =62) | 42.48 (18-78) | Reactive (n =34) Non-reactive (n =28) |
Reactive (n =34) Non-reactive (n =28) |
Reactive (n =15) Non-reactive (n =47) |
Reactive (n =27) Non-reactive (n =35) |
| Children (n =8) | 13.25 (6-17) | Reactive (n =3 ) Non-reactive (n =5) |
Reactive (n =4) Non-reactive (n =4) |
Reactive (n =3) Non-reactive (n =5) |
Reactive (n =4) Non-reactive (n =4) |
| Total (n =200) | 41.54 | Senstivity = 67% Specificity = 87% |
Senstivity = 72% Specificity = 82% |
||
Figure 1: Reactivity of Saliva and Nasopharyngeal Testing.
Furthermore, the same 200 patients’ nasopharyngeal specimen was used for RT-PCR and antigen rapid test. Now out of 200 patients, 100 showed positive results on RT-PCR i.e mean Ct value less than 40 and 81 (50 males, 27 females, and 4 children) showed reactivity i.e positive results on nasal-based antigen rapid test kits. In comparison with RT-PCR and antigen rapid test kit for saliva, showed sensitivity and specificity of 72% and 82% respectively, as shown in Table 1.
These sensitivities of our lollipop style SARS-CoV-2 Antigen rapid test kits are not appreciable and so these rapid diagnostic kits cannot be used as a reliable and accurate screening kit.
The COVID-19 monitoring and diagnosis of SARS-CoV-2 are significant public health issues in third-world nations with severe socioeconomic gaps and inferior healthcare systems. According to WHO recommendations, the SARS-CoV-2 RDTs testing should have a minimum sensitivity of ≥ 80% and specificity of ≥ 97% [18].
Since the same sample materials were used to compare RT-PCR to RDTs rather than utilizing distinct specimens, there was no chance for distribution error, which is a benefit of this study. Because SAR-CoV-2 replication is greater in the pharynx in the early days following infection and then diminishes [16,25]. It might be the reason, that the sensitivity of nasopharyngeal swab-based antigen tests is high during the early stages of infection. Considering that the findings of both rapid tests were unsatisfactory, a combination-test strategy is also recommended for reliable COVID-19 diagnosis.
The number of viruses is growing, thus innovative molecular techniques should be investigated to consider signal transduction pathways and potential host proteins that affect viral replication (Saeed U, Piracha ZZ) [26-30]. The results extracted in the current study are crucial for national strategic organizations that make policy decisions, but it also satisfies a global need for precise COVID-19 diagnostic testing. Using the findings of this study, improved testing procedures may be developed that would solve technical and economical problems. Executing the COVID-19 RDT testing approach involves several hurdles, such as developing policies, finding qualified staff, creating quality assurance standards, and addressing technical challenges. Hence, it is important to regularly assess RDT-based COVID-19 kits among Pakistani populations. Before marketing, the COVID-19 RDT kits, proper usage, and quality must be assured. Furthermore, it is strongly advised that the government ensure the adoption of standard operating procedures for the validation of national testing methods regularly throughout time.
Rapid and precise detection of COVID-19 is needed to avoid the worst health scenarios. In developing countries like Pakistan having a delicate health care system, poor quality RDT kits can cause severe health issues. The study investigated that the sensitivity and specificity of saliva-based kits are 67% and 87% respectively, and that of NPS-RDT is 72% and 82% respectively, which need further research and improvements.
We would like to acknowledge the International Center of Medical Sciences Research (ICMSR), Islamabad Pakistan for providing research facilities, funding, and technical support for manuscript writing and publishing.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579(7798):270-273. doi: 10.1038/s41586-020-2012-7. Epub 2020 Feb 3. PMID: 32015507; PMCID: PMC7095418.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-269. doi: 10.1038/s41586-020-2008-3. Epub 2020 Feb 3. Erratum in: Nature. 2020 Apr;580(7803):E7. PMID: 32015508; PMCID: PMC7094943.
- World Health Organization Coronavirus Disease 2019 (COVID-19) Situation Report-97. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200426-sitrep-97-covid-19.pdf
- Ciotti M, Ciccozzi M, Terrinoni A, Jiang WC, Wang CB, Bernardini S. The COVID-19 pandemic. Crit Rev Clin Lab Sci. 2020 Sep;57(6):365-388. doi: 10.1080/10408363.2020.1783198. Epub 2020 Jul 9. PMID: 32645276.
- Worldometers COVID-19. Available at https://www.worldometers.info/coronavirus/
- WHO Coronavirus disease 2019 COVID-19 Situation report - 184 2020. World Health Organization Accessed July 23, 2020.
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020 Apr;26(4):450-452. doi: 10.1038/s41591-020-0820-9. PMID: 32284615; PMCID: PMC7095063.
- Nao N, Yamagishi J, Miyamoto H, Igarashi M, Manzoor R, Ohnuma A, Tsuda Y, Furuyama W, Shigeno A, Kajihara M, Kishida N, Yoshida R, Takada A. Genetic Predisposition To Acquire a Polybasic Cleavage Site for Highly Pathogenic Avian Influenza Virus Hemagglutinin. mBio. 2017 Feb 14;8(1):e02298-16. doi: 10.1128/mBio.02298-16. PMID: 28196963; PMCID: PMC5312086.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-269. doi: 10.1038/s41586-020-2008-3. Epub 2020 Feb 3. Erratum in: Nature. 2020 Apr;580(7803):E7. PMID: 32015508; PMCID: PMC7094943.
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb 22;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8. Epub 2020 Jan 30. PMID: 32007145; PMCID: PMC7159086.
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI; Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020 Aug 20;182(4):812-827.e19. doi: 10.1016/j.cell.2020.06.043. Epub 2020 Jul 3. PMID: 32697968; PMCID: PMC7332439.
- Kumar N, Shahul Hameed SK, Babu GR, Venkataswamy MM, Dinesh P, Kumar Bg P, John DA, Desai A, Ravi V. Descriptive epidemiology of SARS-CoV-2 infection in Karnataka state, South India: Transmission dynamics of symptomatic vs. asymptomatic infections. EClinicalMedicine. 2021 Feb;32:100717. doi: 10.1016/j.eclinm.2020.100717. Epub 2021 Jan 6. PMID: 33521608; PMCID: PMC7831811.
- Procop GW, Brock JE, Reineks EZ, Shrestha NK, Demkowicz R, Cook E, Ababneh E, Harrington SM. A Comparison of Five SARS-CoV-2 Molecular Assays With Clinical Correlations. Am J Clin Pathol. 2021 Jan 4;155(1):69-78. doi: 10.1093/ajcp/aqaa181. PMID: 33015712; PMCID: PMC7665304.
- Garg A, Ghoshal U, Patel SS, Singh DV, Arya AK, Vasanth S, Pandey A, Srivastava N. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. 2021 Apr;93(4):2281-2286. doi: 10.1002/jmv.26691. Epub 2020 Dec 17. PMID: 33230819; PMCID: PMC7753435.
- Ezzikouri S, Nourlil J, Benjelloun S, Kohara M, Tsukiyama-Kohara K. Coronavirus disease 2019-Historical context, virology, pathogenesis, immunotherapy, and vaccine development. Hum Vaccin Immunother. 2020 Dec 1;16(12):2992-3000. doi: 10.1080/21645515.2020.1787068. Epub 2020 Aug 5. PMID: 32755425; PMCID: PMC8641599.
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020 Mar 19;382(12):1177-1179. doi: 10.1056/NEJMc2001737. Epub 2020 Feb 19. PMID: 32074444; PMCID: PMC7121626.
- Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020 Jun 25;58(7):1070-1076. doi: 10.1515/cclm-2020-0285. PMID: 32172228.
- WHO. Laboratory biosafety guidance related to the novel coronavirus (2019-nCoV). Interim guidance. 2020. Available at https://www.who.int/docs/default-source/coronavir-use/laboratory-biosafety-novel-coronavirus-version-1-1.pdf?
- Pallett SJC, Rayment M, Patel A, Fitzgerald-Smith SAM, Denny SJ, Charani E, Mai AL, Gilmour KC, Hatcher J, Scott C, Randell P, Mughal N, Jones R, Moore LSP, Davies GW. Point-of-care serological assays for delayed SARS-CoV-2 case identification among health-care workers in the UK: a prospective multicentre cohort study. Lancet Respir Med. 2020 Sep;8(9):885-894. doi: 10.1016/S2213-2600(20)30315-5. Epub 2020 Jul 24. Erratum in: Lancet Respir Med. 2020 Jul 30;: PMID: 32717210; PMCID: PMC7380925.
- Xu R, Cui B, Duan X, Zhang P, Zhou X, Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020 Apr 17;12(1):11. doi: 10.1038/s41368-020-0080-z. PMID: 32300101; PMCID: PMC7162686.
- Fakheran O, Dehghannejad M, Khademi A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect Dis Poverty. 2020 Jul 22;9(1):100. doi: 10.1186/s40249-020-00728-w. PMID: 32698862; PMCID: PMC7374661.
- To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, Leung WS, Chik TS, Choi CY, Kandamby DH, Lung DC, Tam AR, Poon RW, Fung AY, Hung IF, Cheng VC, Chan JF, Yuen KY. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin Infect Dis. 2020 Jul 28;71(15):841-843. doi: 10.1093/cid/ciaa149. PMID: 32047895; PMCID: PMC7108139.
- Fakheran O, Dehghannejad M, Khademi A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect Dis Poverty. 2020 Jul 22;9(1):100. doi: 10.1186/s40249-020-00728-w. PMID: 32698862; PMCID: PMC7374661.
- Saeed U, Kim J, Piracha ZZ, Kwon H, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner. J Virol. 2019 Mar 5;93(6):e01840-18. doi: 10.1128/JVI.01840-18. PMID: 30567987; PMCID: PMC6401437.
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 May;581(7809):465-469. doi: 10.1038/s41586-020-2196-x. Epub 2020 Apr 1. Erratum in: Nature. 2020 Dec;588(7839):E35. PMID: 32235945.
- Piracha ZZ, Kwon H, Saeed U, Kim J, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. Sirtuin 2 Isoform 1 Enhances Hepatitis B Virus RNA Transcription and DNA Synthesis through the AKT/GSK-3β/β-Catenin Signaling Pathway. J Virol. 2018 Oct 12;92(21):e00955-18. doi: 10.1128/JVI.00955-18. PMID: 30111572; PMCID: PMC6189494.
- Piracha ZZ, Saeed U, Kim J, Kwon H, Chwae YJ, Lee HW, Lim JH, Park S, Shin HJ, Kim K. An Alternatively Spliced Sirtuin 2 Isoform 5 Inhibits Hepatitis B Virus Replication from cccDNA by Repressing Epigenetic Modifications Made by Histone Lysine Methyltransferases. J Virol. 2020 Jul 30;94(16):e00926-20. doi: 10.1128/JVI.00926-20. PMID: 32493816; PMCID: PMC7394897.
- Saeed U, Piracha ZZ. Viral outbreaks and communicable health hazards due to devastating foods in Pakistan.World J Virol. 2016 May 12;5(2):82–4.
- Saeed U, Piracha ZZ, Manzoor S. Hepatitis C virus induces oxidative stress and DNA damage by regulating DNAPKCs, ATM, ATR and PARP mediated signaling and guards cell from cancerous condition by upregulating RB, P53 and downregulating VEGF. Acta Virol. 2017;61(3):316–23.
- Saeed U, Uppal SR, Piracha ZZ, Rasheed A, Aftab Z, Zaheer H, Uppal R. Evaluation of SARS-CoV-2 antigen-based rapid diagnostic kits in Pakistan: formulation of COVID-19 national testing strategy. Virol J. 2021 Feb 13;18(1):34.