More Information
Submitted: July 30, 2024 | Approved: August 12, 2024 | Published: August 13, 2024
How to cite this article: RICHARD R, EZEJIOFOR T.I.N, NSOFOR C.A, MANINGI N.E. Antibacterial Resistance and Extended-Spectrum Beta-Lactamase (ESBL) Phenotypes in Enterobacteriaceae Isolated from Fecal Samples of Humans and Animals in Selected Local Government Areas of Nasarawa State, Nigeria. Arch Biotechnol Biomed. 2024; 8(1): 027-033. Available from: https://dx.doi.org/10.29328/journal.abb.1001041.
DOI: 10.29328/journal.abb.1001041
Copyright License: © 2024 RICHARD R, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Enterobacteriaceae; Phenotypic; Antibacterial; Resistance; Occurrence
Abbreviations: ESBL: Extended Spectrum Beta Lactamase; CLSI: Clinical and Laboratory Standards Institute; MAR: Multiple Antibiotics Resistance; MDR: Multi-Drug Resistant; PAN: Pan Drug Resistant; XDR: Extensive Drug Resistant; UTI: Urinary Tract Infections; DDST: Double Disc Synergy Test; LGA: Local Government Area
Antibacterial Resistance and Extended-Spectrum Beta-Lactamase (ESBL) Phenotypes in Enterobacteriaceae Isolated from Fecal Samples of Humans and Animals in Selected Local Government Areas of Nasarawa State, Nigeria
RICHARD R1*, EZEJIOFOR T.I.N2, NSOFOR C.A2 and MANINGI N.E3
1Department of Biological Science, Federal University Gashua, P.M.B. 1005, Yobe state, Nigeria
2Department of Biotechnology, Federal University of Technology, Owerri, P.M.B. 1526, Imo state, Nigeria
3Department of Microbiology, School of Life Sciences, University of Kwazulu Natal, South Africa
*Address for Correspondence: RICHARD R, Department of Biological Science, Federal, University Gashua, P.M.B. 1005, Yobe state, Nigeria, Email: [email protected]
It is quite alarming the increasing rate of antibacterial resistance all over the world considering the public health threat and the re-emergence of multi-drug resistant Enterobacteriaceae. The aim of this study is Antibacterial resistance and phenotypic detection of Extended Spectrum Beta-Lactamase (ESBL) producing Enterobacteriaceae isolated from human and animal fecal samples in selected local government areas of Nasarawa state, Nigeria was carried out in the study. Hundred (100) samples comprising human and animal (goats, cattle, and chicken) were collected and 55 samples were multidrug resistant. A commercial biochemical kit (Eneterosystem 18R) was used for the isolation and identification of Enterobacteriaceae. Kirby Bauer Disk Diffusion Method was used for antibacterial susceptibility testing of Enterobacteriaceae isolates. The Double Disc Synergy Test (DDST) method was also used for the phenotypic confirmation test of Extended Spectrum Beta Lactamase (ESBL). The occurrence of multidrug-resistant isolates shows that Escherichia coli (100.00%) which is the highest, Proteus mirabilis (14.54%), Klebsiella pneumoniae, and Salmonella enterica (10.90%), while the occurrence of Shigella flexneri (9.09%) was the lowest. The Enterobacteriaceae isolates were more resistant to Cefuroxime, Cefexime, Amoxicillin Clavulanate, and Imipenem/Cilastatin with percentage resistance ranges from 66.6% - 100%. The occurrence of ESBL producers shows that Escherichia coli (60.00%) and Proteus mirabilis (62.5%) were high while Shigella flexneri (20.0%) had a low occurrence of ESBL. The sale and in-discriminate use of antibiotics without a prescription is an important regulatory issue in the abuse of antibiotics for both humans and animals. The Beta-Lactam and gentamycin antibiotics were not effective against the Multi-Drug Resistant (MDR) isolates and most of the isolates were ESBL producers.
Antimicrobial Resistance (AMR) has been reported by Gonzales-Rodriguez, et al. [1] to be a serious threat to public health. All antibiotics will become ineffective by 2050 as estimated by the World Health Organization (WHO) which could be due to misuse or inappropriate use of antibiotics [1].
Antimicrobial-Resistant Bacteria (ARB) and Antimicrobial Resistance Genes (ARGs) mostly find their way into the environment through diverse means such as feces, urine, and milk [2], which may spread to humans either directly or indirectly through the food chain that is milk and meat or manure land application as soil amendment for vegetables and crops for human consumption [2,3].
In animal and human health care, antimicrobials have been used which has increased the widespread occurrence of antimicrobial-resistant bacteria not just in animals and humans, but more so in the environment such as surface water, air, and soil. It is obvious that the possibility of acquiring antimicrobial-resistant bacteria outside health care has increased, for example, individuals may acquire it through the preparation and consumption of meat products that are contaminated, herbs, fruits, and vegetables, or even contact with or the ingestion of contaminated surface water, for example during recreational activities [4-6].
Enterobacteriaceae have been reported by Jude, et al. [7] to be a serious health threat to the public because of the different resistance mechanisms of Enterobacteriaceae and under production of new drugs which have made microorganisms resistant to almost all available antibiotics.
Extended-spectrum Beta-lactamase (ESBL) is an enzyme produced by Enterobacteriaceae (Gram-negative bacteria), which is a great threat to health in the fields of human and veterinary medicine Worldwide [8].
There is an increase in MDR Enterobacteriaceae Worldwide which is quite alarming considering the threat to public health, series of research of late have reported the emergence of MDR bacterial pathogen from different sources including soil, air, flowing rivers, vegetables, wastewater, refuse dump sites, human and animals [9,10].
The aim of this study was to investigate Antibacterial Resistance and Phenotypic Detection of Extended Spectrum Beta-Lactamase (ESBL) Producing Enterobacteriaceae Isolated from Fecal Sample of Human and Animal (Goats, Cattle, and Chicken) in selected local government areas of Nasarawa State, Nigeria.
Materials
The antibiotic discs used for susceptibility testing includes: Amoxicillin-Clavulanic acid (AUG: 30 μg), Cefotaxime (CTX: 25 μg), Imipenem/Cilastatin (IMP: 10/10 μg), Ofloxacin (OFX: 5 μg), Gentamicin (GN: 10 μg), Nalidixic Acid (NA: 30 μg), Cefuroxime (CXM: 30 μg), Ceftriaxone Sulbactarm (CRO: 45 μg), Cefexime (ZEM: 5 μg) and Levofloxacin (LBC: 5 μg). All were products of Celtech Diagnostic Belgium.
The antibiotic discs used for phenotypic detection in this study include Amoxicillin-Clavulanic acid (AMT: 30 μg), Cefotaxime (CTX: 30 μg), Ceftazidime (CAZ: 30 μg). All discs were products of Oxoid Ltd (U.K.).
Enterosystem 18R kits were used for the identification of Enterobacteriaceae (Liofilchem Diagnostic Italy).
Ethical approval
Ethical approval was obtained from the Nasarawa State Ministry of Health, Nasarawa State with REG. NO: NHREC 18/08/2017, the approval letter was dated 29th November 2021.
Methods
Hundred (100) fecal samples were collected in Keffi, Karu, Akwanga, and Lafia local government areas of Nasarawa State.
Sampling techniques
Human fecal sample: Human fecal samples of patients that were submitted to the microbiology laboratory for normal routine services were collected over a period of three months using a sterile container and transported using an ice pack within 6 hours to the Microbiology Laboratory, Nasarawa State University, keffi for analysis.
Enterobacteriaceae were isolated from fecal samples of patients with the aid of a sterile wire loop the fecal samples were streaked on MacConkey agar and or Eosin Methylene Blue (E.M.B) agar. The plates were incubated at 37 °C for 24 hours - 48 hours [11]. After which, presumptive Enterobacteriaceae colonies were sub-cultured on nutrient agar and incubated at 37 °C for 24 hours - 48 hours, those that showed positive growth were Gram stained and viewed under the microscope using oil immersion x100 objectives lens [9,12].
Animal fecal sample: Fresh fecal droppings from goats, cattle, and chickens were randomly collected using sterile containers and care was taken to avoid collecting more than one fecal sample per individual animal [13]. The animal fecal samples were transported using an ice pack within 6 hours to the Microbiology Laboratory, Nasarawa State University, Keffi for analysis.
Enterobacteriaceae were isolated from fecal samples of animals with the aid of a sterile wire loop the fecal samples were streaked on MacConkey agar and or Eosin Methylene Blue (E.M.B) agar. The plates were incubated at 37 °C for 24 hours - 48 hours. After which, presumptive Enterobacteriaceae colonies were sub-cultured on nutrient agar and incubated at 37 °C for 24 hours - 48 hours, those that showed positive growth were Gram stained and viewed under the microscope using oil immersion x100 objectives lens [9,12].
Identification and biochemical characterization of Enterobacteriaceae isolated from fecal samples of human and animal in selected local government areas of Nasarawa state, Nigeria
The presumptive Enterobacteriaceae isolates were identified by microscopy (gram staining) and some basic biochemical characterization tests were carried out which included; a citrate test, indole test, and Methyl red- Voges Proskauer was chosen according to Bergy, manual of determinative bacteriology by Holt, et al. [12]; Cheesbrough, [14] and they were further identified using Commercial Biochemical Kit (Enterosystem 18R). In summary, the presumptive Enterobacteriaceae to be identified was recently isolated (18 h – 24 h); one or more morphologically similar well-isolated colonies from the agar culture medium were suspended in a physiological solution and the suspension was thoroughly homogenized [9].
The system was unwrapped and brought to room temperature, the system was labelled properly, Bacterial suspension of about 0.2 ml was transferred into each well of the system and overlaid with 1 drop Vaseline oil in the wells 2-LDC, 3-ODC, 4-ADC, 7-UR, and 8-H2S. The system was then covered with a lid provided and incubated at 36 ± 1 OC for 12-18-24 hours [9].
After the incubation period, 2 drops of alpha-napthol and 1 drop of NaOH 40% was added to the well 10-VP presence of pink-red colour in about 20 minutes indicates positive reaction, about 2-3 drops of KOVAC’S Reagents was added into the well 11-IND, the presence of red colour within 2-3 minutes indicates positive reaction, colour change was watched at each well and the results were carefully read and interpreted using the standard given in the identification index [9].
Antibacterial susceptibility test of Enterobacteriaceae isolated from fecal sample of human and animal in selected local government areas of Nasarawa state, Nigeria
The antibacterial susceptibility test of the bacterial isolates was carried out as earlier described by the Clinical and Laboratory Standards Institute [15]. Briefly, three (3) pure colonies of the isolates were inoculated into 5 ml sterile 0.85% (w/v) NaCl (normal saline), and the turbidity of the bacteria suspension was adjusted to the turbidity equivalent to 0.5 McFarland’s standard. The McFarland’s standard was prepared as follows: 0.5 ml of 1.172% (w/v) BaCl2.2H2O (BDH Chemicals Ltd England) was added into 99.5 ml of 1% (w/v) H2SO4 (BDH Chemicals Ltd England).
A sterile swab stick was soaked in the bacteria suspension and streaked on Mueller-Hinton agar (Oxoid Ltd., Basingstoke, UK) plates, the antibiotic discs were placed at the center of the petri plates aseptically and allowed to stand for 1 h for pre-diffusion. The petri plates were incubated at 37 OC for 24 h. The diameter zone of inhibition in millimeters was measured using a ruler and the result was interpreted in accordance with the susceptibility breakpoint earlier described by the Clinical and Laboratory Standards Institute [15].
Determination of Multiple Antibiotic Resistance (MAR) index of Enterobacteriaceae isolated from fecal sample of human and animal in selected local government areas of Nasarawa state, Nigeria
The MAR index of the Enterobacteriaceae isolates was determined using the method of Krumperman [16] with little adjustments by Tsaku, et al. [17]: MAR Index = No. antibiotics isolate is resistant to/No. of antibiotics tested.
Antibiotic resistance classification
Antibiotic resistance in the Enterobacteriaceae isolates was classified into: multidrug-resistant; extensive drug-resistant and pan drug-resistant [18].
Phenotypic confirmatory test for ESBL production of Enterobacteriaceae isolated from fecal sample of human and animal in selected local government areas of Nasarawa state, Nigeria
Phenotypic confirmatory test of ESBL production by Enterobacteriaceae isolates resistant to both third and generation Cephalosporins (Ceftazidime and Cefotaxime) was carried out using the Double Disc Synergy Test (DDST) method as described earlier by Jarlier, et al. [19]. In brief, 105cfu/ml bacterial suspension was streaked on sterile Mueller-Hinton agar plates and the Amoxicillin-Clavulanic acid (AMT: 30 μg) disc was placed at the center of the plate. Cefotaxime (CTX: 30 μg) and Ceftazidime (CAZ: 30 μg) discs were placed at 15 mm (edge to edge) from the center disc. The enhancement of the zone of inhibition in the area between the Amoxicillin-Clavulanic acid disc and any one of the β-lactam discs compared with the zone of inhibition on the far side of the drugs disc was interpreted as the presence of an ESBL in the test strain.
1. Cultural, morphological, and biochemical characteriza-tion of Enterobacteriaceae isolated from fecal samples of humans and animals in selected local government areas of Nasarawa state, Nigeria
The cultural, morphological, and biochemical characterization of several species of enterobacteriaceae isolated from human and animal fecal samples include; Escherichia coli, Proteus mirabilis, Salmonella enterica, Klebsiella pneumonia, and Shigella flexneri as shown in Table 1.
| Table 1: Cultural, Morphological, and Biochemical Characterization of Enterobacteriaceae Isolated from Fecal Samples of Humans and Animals in Selected Local Government Areas of Nasarawa State, Nigeria. | |||||||||||||||||||||
| Culture characteristics | Morphology characteristics | Biochemical characteristics | Inference | ||||||||||||||||||
| Gram reaction | Morphology | ONPG | LDC | ODC | ADC | PD | CIT | UR | H2S | MLN | VP | IND | GLU | MAN | INO | SOR | SAC | ARA | |||
| Pinkish colonies on MAC and greenish metallic sheen on EMB agar. | - | Rod | + | + | V | V | _ | _ | _ | _ | _ | _ | + | + | + | _ | V | V | V | V | Escherichia coli |
| Pale or colourless colonies on MCA and it gives a strong fishy odour | _ | Rod | _ | _ | V | _ | V | V | + | V | _ | _ | V | + | _ | _ | _ | V | _ | _ | Proteus mirabilis. |
| Blackish colonies on Salmonella shigella agar (SSA) | _ | Rod | _ | + | + | V | _ | V | _ | V | _ | _ | _ | + | + | V | V | _ | V | _ | Salmonella enterica. |
| Pink on MCA. Pink-purple colonies with no metallic sheen on EMB agar | _ | Rod | + | V | _ | _ | _ | + | V | _ | V | V | _ | + | + | V | + | + | + | + | Klebsiella pneumoniae. |
| Milkish/ colourless on SSA with a black center | _ | Rod | V | _ | _ | V | _ | _ | _ | _ | _ | _ | V | + | V | _ | V | _ | V | V | Shigella flexneri |
| KEYS: + = positive; - = negative V = variable (10-95% positively)ONPG = Hydrolysis of ONPG (Ortho-Nitrophenyl-β-Galactoside) LDC: Decarboxylation of Lysine; ODC: Decarboxylation of Ornithine; ADC: Decarboxylation of Arginine; PD” Decarboxylation of Phenylalanine; CIT: Utilization of Citrate; UR: Hydrolysis of Urea; H2S: Production of Hydrogen Sulphide; MLN: Utilization of Malonate; VP: Production of Acetoin (Voges-Proskauer test); IND: Production of Indole (Kovacs reagent); GLU: Fermentation of Glucose; MAN: Fermentation of Mannitol; INO: Fermentation of Inositol; SOR: Fermentation of Sorbitol; SAC: Fermentation of Saccharose; ARA: Fermentation of Arabinose; RAF: Fermentation of Raffinose |
|||||||||||||||||||||
2. Occurrence of Enterobacteriaceae isolated from fecal samples of humans and animals in selected local government areas of Nasarawa state, Nigeria
The occurrence of Enterobacteriaceae isolated from human and animal fecal samples in selected local government areas of Nasarawa state, Nigeria is shown in Table 2. Out of 55 MDR human and animal fecal samples, the occurrence of Escherichia coli (100.0%) was the highest while the occurrence of Shigella flexneri (9.09%) was the lowest.
| Table 2: The Occurrence of Multi-Drug Resistant Enterobacteriaceae isolated from Human and Animal Fecal Samples in Selected Local Government Areas of Nasarawa State, Nigeria. | ||||||
| Fecal Samples | No of MDR | No. (%) Enterobacteriaceae | ||||
| ES | PR | SA | KL | SH | ||
| Human | 30 | 30(100.0) | 5(16.5%) | 2(6.6%) | 4(13.3%) | 3(10.0%) |
| Animal | 25 | 25(100.0) | 3(12.0%) | 4(16.0%) | 2(8.0%) | 2(8.0%) |
| Total | 55 | 55(100.0) | 8(14.54%) | 6(10.90%) | 6(10.90%) | 5(9.09%) |
| Keys: MDR: Multidrug Resistant; ES: Escherichia coli; PR: Proteus mirabilis; SA: Salmonella enteric; KL: Klebsiella pneumonia; SH: Shigella flexneri | ||||||
3. Antimicrobial resistance of Enterobacteriaceae isolated from fecal samples of humans and animals in selected local government areas of Nasarawa state, Nigeria
Antimicrobial resistance of Enterobacteriaceae isolated from human and animal samples in selected local government areas of Nasarawa state Nigeria is as shown in Table 3. The Escherichia coli, Proteus mirabilis, Salmonella enterica, Klebsiella pneumoniae, and Shigella flexneri were resistant to Amoxicillin Clavulanic acid, Cefexime, Cefuroxime and Ceftriaxone Sulbactarm with resistance ranges from 66.6% - 100% but less resistance to levofloxacin and ofloxacin with percentage resistance ranges from 12.5% - 50.0% respectively as shown in Table 3.
| Table 3: The Antimicrobial Resistance Patterns of Enterobacteriaceae Isolated from Human and Animal Fecal in Nasarawa State, Nigeria. | ||||||
| Antimicrobials | Disc Contents (µg) | No. (%) Resistance | ||||
| ES (N = 55) |
PR (N = 8) |
SA (N = 6) |
KL (N = 6) |
SH (N = 5) |
||
| CXM | 30 | 53(96.36) | 7(87.5) | 4(66.6) | 6(100.0) | 5(100.0) |
| CTX | 25 | 47(85.45) | 6(75.0) | 3(50.0) | 5(83.8) | 5(100.0) |
| IMP | 10/10 | 55(100.0) | 8(100.0) | 6(100.0) | 4(66.6) | 5(100.0) |
| OFX | 5 | 13(23.63) | 4(50.0) | 2(33.3) | 1(16.6) | 1(20.0) |
| GN | 10 | 10(18.18) | 3(37.5) | 2(33.3) | 3(50.0) | 2(40.0) |
| NA | 30 | 54(98.18) | 7(87.5) | 3(50.0) | 3(50.0) | 3(60.0) |
| LBC | 5 | 8(14.54) | 1(12.5) | 1(16.6) | 2(33.3) | 2(40.0) |
| CRO | 45 | 53(96.36) | 7(87.5) | 5(83.8) | 4(66.6) | 4(80.0) |
| AMC | 30 | 55(100.0) | 8(100.0) | 6(100.0) | 6(100.0) | 5(100.0) |
| ZEM | 5 | 55(100.0) | 8(100.0) | 5(83.8) | 6(100.0) | 5(100.0) |
| Keys: CXM: Cefuroxime; CTX: Cefotaxime; IMP: Imipenem/Cilastatin; OFX: Ofloxacin; GN: Gentamycin; NA: Nalidixic Acid; LBC: Levofloxacin; CRO: Ceftriaxone Sulbactarm; AMC: Amoxicillin Clavulanate; ZEM: Cefexime; MDR: Multidrug resistant; ES: Escherichia coli; PR: Proteus mirabilis; SA: Salmonella enteric; KL: Klebsiella pneumonia; SH: Shigella flexneri | ||||||
4. Extended-spectrum beta-lactamase production of Enterobacteriaceae isolated from fecal samples of humans and animals in selected local government areas of Nasarawa state, Nigeria
The production of ESBL by different species of MDR Enterobacteriaceae isolated from human and animal samples is shown in Table 4. The occurrence of ESBL producers was high in Proteus mirabilis (62.5%), Escherichia coli (60.0%), and Klebsiella pneumoniae (50.0%) but low in Salmonella enterica (33.3%) and Shigella flexneri (20.0%) respectively as shown in Table 4.
| Table 4: Extended Spectrum Beta Lactamase phenotypes observed in Enterobacteriaceae Isolated from Human and Animal Fecal in Nasarawa State, Nigeria. | |||||
| ESBL Production | No. (%) Enterobacteriaceae | ||||
| ES (N = 55) |
PR (N = 8) |
SA (N = 6) |
KL (N = 6) |
SH (N = 5) |
|
| POSITIVE | 33(60.0) | 5(62.5) | 2(33.3) | 3(50.0) | 1(20.0) |
| NEGATIVE | 22(40.0) | 3(37.5) | 4(66.4) | 3(50.0) | 4(80.0) |
| Keys: MDR: Multi-Drug Resistant; XDR: Extensive Drug Resistant; PAN: Pan Drug Resistant; ES: Escherichia coli; PR: Proteus mirabilis;SA: Salmonella enterica; KL: Klebsiella pneumonia; SH: Shigella flexneri | |||||
5. Categories of antimicrobial resistance of Enterobacteriaceae isolated from fecal samples of humans and animals in selected local government areas of Nasarawa state, Nigeria
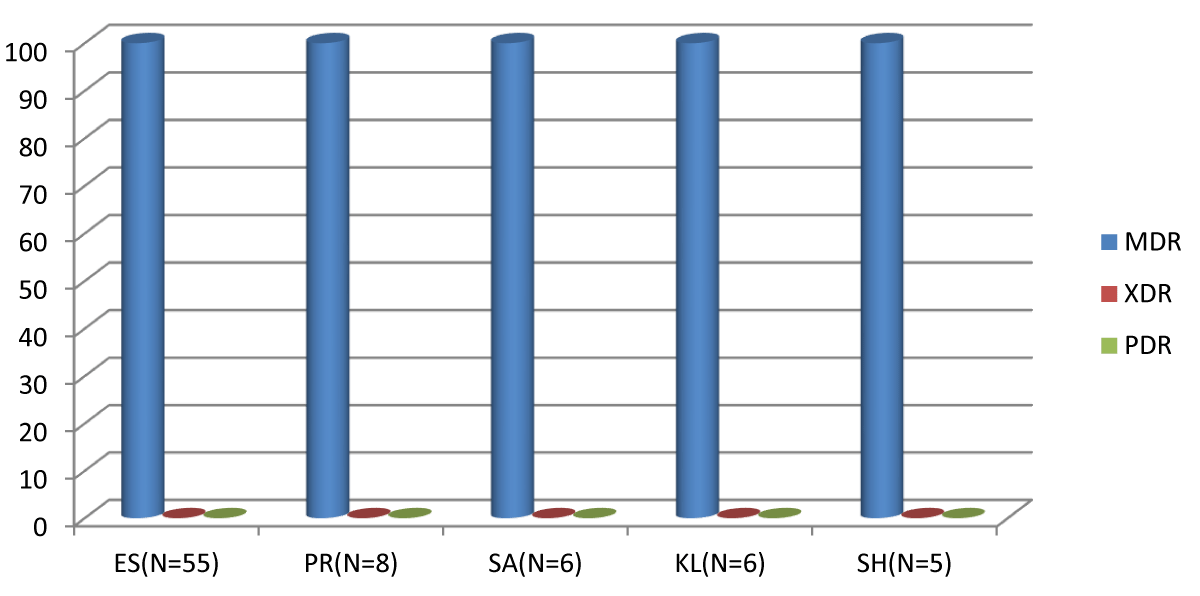
The antimicrobial-resistant Enterobacteria isolated from human and animal samples in selected local government areas of Nasarawa state, Nigeria was classified into; Multi-Drug Resistant (MDR), extensive drug-resistant (XDR), and pan drug-resistant (PAN) with occurrence of MDR as the highest with percentage occurrence of 100% but none of the antimicrobial resistant enterobacteriacea were either extensive or pan drug resistance as shown in Figure 1.
Figure 1: The Classes of Antimicrobial Resistance in Antimicrobial Resistant of Enterobacteriaceae Isolated from Human and Animal Fecal in Nasarawa State, Nigeria. Keys: MDR: Multi-Drug Resistant; XDR: Extensive Drug Resistant; PAN: Pan Drug Resistant; ES: Escherichia coli; PR: Proteus mirabilis; SA: Salmonella enterica; KL: Klebsiella pneumonia; SH: Shigella flexneri
In this study, Escherichia coli, Proteus mirabilis, Salmonella enterica, Klebsiella pneumonia, and Shigella flexneri were isolated from human and animal fecal samples in selected local government areas of Nasarawa state, Nigeria.
MDR Escherichia coli (100%) was isolated from both human and animal fecal samples which is in agreement with the findings of Kristianingtyas, et al. [8]; Tama, et al. [20]; Abimiku, et al. [21]; Ali, et al. [22] and Adebola, et al. [23], but contrary to the findings of Ibrahim, et al. [24] (53.4%) and Langata, et al. [25] (57%). The detection of Escherichia coli at a high rate in this study could be a result of improper hygiene.
The isolates which include; Escherichia coli, Proteus mirabilis, Salmonella enterica, Klebsiella pneumoniae, and Shigella flexneri were resistant to Cefuroxime, Ceftriaxone Sulbactarm, Amoxicillin Clavulanic acid, and Cefexime, with the resistance ranging from 66.6 -100% which is in line with the findings of Barns, et al. [26] which has 100% for each Amoxicillin Clavulanic acid, Cefexime, Ceftazidime, and Cefuroxime respectively but contrary to the study of Tama, et al. [20] Amoxicillin Clavulanic acid (61.1%), Cefuroxime (31.1%), Ceftazime (35.6%) and Imipenem (22.2%), but less resistance to Ofloxacin and Levofloxacin with percentage resistance ranging from 12.5% - 50.0% respectively which is not in agreement with the findings of Barns, et al. [26] Ofloxacin (88.23%). The sale and use of medicines (antibiotics) without a proper prescription is an important regulatory issue in the abuse/ misuse of antibiotics for both humans and animals. The low prevalence rate of fluoroquinolone-resistant enterobacteria is an indication that it has not been abused in the study area and the limited use of fluoroquinolone antibiotics for preventive measures. The extensive use of beta-lactamase and fluoroquinolone antibiotics in human and animal medicine is associated with the increasing emergence of ESBL and quinolone-resistant strains [27]. Several reasons have been attributed to the widespread, of which high levels of beta-lactamase and quinolones use in human and animal (veterinary) medicine is considered to be an important factor [27,28]. Researchers have reported that a factor of concern associated with the advent of quinolone resistance is their close relation with other agents, especially the expression of Extended Spectrum Beta Lactamases (ESBLs) and aminoglycosides [27,29]. It is quite unfortunate that this biological relationship among these agents has caused a suitable opportunity for the dissemination of multidrug resistance among Enterobacteriaceae, thus resulting in restrictions on treatment choices. Therefore, this should be of concern to physicians when prescribing quinolones that the resistance to cephalosporins and aminoglycosides and other resistance forms that are associated with PMQRs may occur as well [27,30].
The high rates of resistance found in this study could be explained by the wide use of antibiotics in Nigeria for prophylaxis and treatments in both humans and animals. The high rate of antibiotic utilization is a great factor in the emergence and dissemination of antibiotic-resistant Enterobacteriaceae including ESBL producers [20]. The trend of antimicrobial resistance among Enterobacteriaceae in food animals such as chickens is a cause of concern, especially due to the possibility and potential for the transfer of these pathogens to the human population [20]. An increase in the spread of ESBL-producing bacteria has been recorded due to excessive usage of β-lactams in both humans and animals, which is threatening personnel in the poultry industry and consequently posing a threat to human health [31]. ESBLs are widespread in Enterobacteriaceae, especially in E. coli and Salmonella sp., while ESBL-producing E. coli has been reported in food animals worldwide Tsekouras, et al. [31]. There are reports associated with human ESBL carriage with exposure to Extended Spectrum Beta Lactamase-producing Enterobacteriaceae of livestock origin, raising concerns about the possible transfer of ESBL producers through the food chain, which could jeopardize public health Tsekouras, et al. [31].
The presence of ESBL-producing Enterobacteriaceae in animal samples within the southeastern part of Nigeria has been reported by Chah, et al. [32]. In Owerri, a prevalence rate of 22.2% was reported among poultry [33]. In Sokoto, a prevalence rate of 8.9% and 5.7% for ESBL-producing E. coli in chickens and retail eggs in Sokoto metropolis was reported by Abubakar, et al. [34].
The occurrence of Enterobacteriaceae namely Escherichia coli, Proteus mirabilis, Salmonella enterica, Klebsiella pneumonia, and Shigella flexneri isolated from human and animal fecal samples were highly resistant to β- lactam antibiotics such as cefexime, cefuroxime, ceftriaxone sulbactam, cefotaxime and imipenem/cilastatin but less resistant to levofloxacin and ofloxacin. These antibiotics may be useful for treatments in the study area most of the isolates are MDR Enterobacteriaceae. The sale and in-discriminate use of antibiotics without a prescription is an important regulatory issue in the abuse of antibiotics for both humans and animals.
The authors are thankful to Federal University Gashua Yobe State, Nigeria for creating the avenue for TETFund to fund this research. Also, we are grateful to the Department of Microbiology Nasarawa State University, keffi, for making facilities available for sample collection, analysis, and technical support during the study.
Funding
Funds were made available by TETFund under the training funds allocation through the Federal University Gashua Yobe State, Nigeria.
- Gonzales-Rodriguez A, Reyes-Farias C, Gonzales-Escalante E. Identification of multidrug-resistant Enterobacteriaceae in fecal samples from infants residing in Talara, Piura, Peru. Rev Peru Med Exp Salud Publica. 2022;39(4):456-462. Available from: https://doi.org/10.17843/rpmesp.2022.394.11870
- Gelalcha BD, Ensermu DB, Agga GE, Vancuren M, Gillespie BE, D’Souza DH, et al. Prevalence of antimicrobial resistant and extended-spectrum beta-lactamase-producing Escherichia coli in dairy cattle farms in East Tennessee. Foodborne Pathog Dis. 2022;19(6):408-416. Available from: https://doi.org/10.1089/fpd.2021.0101
- Samreen, Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist. 2021;27:101-111. Available from: https://doi.org/10.1016/j.jgar.2021.08.001
- Blaak H, Lynch G, Italiaander R, Hamidjaja RA, Schets FM, de Roda Husman AM. Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch surface water and wastewater. PLoS One. 2015;10(6). Available from: https://doi.org/10.1371/journal.pone.0127752
- Blaak H, de Kruijf P, Hamidjaja RA, van Hoek AHAM, de Roda Husman AM, Schets FM. Prevalence and characteristics of ESBL-producing E. coli in Dutch recreational waters influenced by wastewater treatment plants. Vet Microbiol. 2014;171:448-459. Available from: https://doi.org/10.1016/j.vetmic.2014.03.007
- Søraas A, Sundsfjord A, Sandven I, Brunborg C, Jenum PA. Risk factors for community-acquired urinary tract infections caused by ESBL-producing Enterobacteriaceae – a case-control study in a low prevalence country. PLoS One. 2013;8. Available from: https://doi.org/10.1371/journal.pone.0069581
- Jude FL, Innocent MA, Ousenu K, Christopher BT. Patterns of antibiotic resistance in Enterobacteriaceae isolates from broiler chicken in West Region of Cameroon: a cross-sectional study. Can J Infect Dis Med Microbiol. 2022;2022:4180336. Available from: https://doi.org/10.1155%2F2022%2F4180336
- Kristianingtyas L, Effendi MH, Witaningrum AM, Wardhana DK, Ugbo EN. Prevalence of extended-spectrum ß-lactamase-producing Escherichia coli in companion dogs in animal clinics, Surabaya, Indonesia. Int J One Health. 2021;7(2):232-236. Available from: https://www.onehealthjournal.org/Vol.7/No.2/12.html.
- Richard R, Ezejiofor TIN, Nsofor CA, Nkene IH. Antibacterial resistance and phenotypic detection of extended spectrum beta-lactamase (ESBL) producing Enterobacteriaceae isolated from environmental sources in Nasarawa State, Nigeria. Eur J Biol Biotechnol. 2023;4(5):612. Available from: https://ejbio.org/index.php/ejbio/article/view/479.
- Jesumirhewe C, Springer B, Allerberger F, Rvopitsch W. Genetic characterization of antibiotic resistant Enterobacteriaceae isolates from bovine animals and environment in Nigeria. Front Microbiol. 2022;13:793541. Available from: https://doi.org/10.3389/fmicb.2022.793541
- Abimiku RH, Ngwai YB, Nkene IH, Bassey BE, Tsaku PA, Ibrahim T, Tama SC, Ishaleku D, Pennap GRI. Molecular diversity and ESBL resistance of diarrheagenic E. coli from patients attending selected health care facilities in Nasarawa State, Nigeria. Int J Pathog Res. 2019;3(1):1-18. Available from: https://doi.org/10.1016/j.heliyon.2019.e02177
- Holt JG, Kieg NR, Sneath PH, Staley JT, Williams ST. Bergey’s Manual of Determinative Bacteriology. 9th ed. Baltimore: Williams & Wilkins. 1994; 786-788.
- Nsofor CA, Iroegbu CU. Plasmid profile of antibiotic resistant Escherichia coli isolated from domestic animals in South-East Nigeria. J Cell Anim Biol. 2013;7(9):109-115. Available from: https://www.researchgate.net/publication/304538332_Journal_of_Cell_and_Animal_Biology_Plasmid_profile_of_antibiotic_resistant_Escherichia_coli_isolated_from_domestic_animals_in_South-East_Nigeria.
- Cheesebrough M. Medical Laboratory Manual for Tropical Countries. Cambridge: Cambridge University Press. 2006; 49-97.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 27th informational supplement M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. Available from: https://clsi.org/media/1469/m100s27_sample.pdf.
- Krumperman PH. Multiple antibiotics indexing E. coli to identifying risk sources of faecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165-170. Available from: https://doi.org/10.1128%2Faem.46.1.165-170.1983
- Tsaku PA, Ngwai YB, Pennap GRI, Ishaleku D, Ibrahim T, Nkene IN, et al. Extended spectrum beta lactamase production of E. coli isolated from door handles in Nasarawa State University, Keffi, Nigeria. Heliyon. 2019;5. Available from: https://doi.org/10.1016%2Fj.heliyon.2019.e02177
- Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-281. Available from: https://doi.org/10.1111/j.1469-0691.2011.03570.x
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867-878. Available from: https://doi.org/10.1093/clinids/10.4.867
- Tama SC, Ngwai YB, Pennap GR, Nkene IH, Abimiku RH, Jodi SM. Antimicrobial resistance profile and extended spectrum beta-lactamase resistance genes in Escherichia coli from poultry droppings Nasarawa, Nigeria. Asian J Biochem Genet Mol Biol. 2021;8(2):47-56. Available from: https://doi.org/10.9734/ajbgmb/2021/v8i230192
- Abimiku RH, Ngwai YB, Nkene IH, Tatfeng YM. Molecular detection of diarrheagenic pathotypes of Escherichia coli from diarrheic patients in Keffi, Nigeria. Microbioz J. 2016;2(3):1-6. Available from: https://microbiozjournals.com/wp-content/uploads/2019/01/REJOICE020316.pdf.
- Ali I, Kumar N, Ahmed S, Dasti JI. Antibiotic resistance in uropathogenic E. coli strains isolated from non-hospitalized patients in Pakistan. J Clin Diagn Res. 2014;8(9). Available from: https://doi.org/10.7860%2FJCDR%2F2014%2F7881.4813
- Adebola O, Oluwatoyin I, Adebayo L. A study of the prevalence of diarrhoeagenic Escherichia coli in children from Gwagwalada, Federal Capital Territory, Nigeria. Pan Afr Med J. 2014;17:146. Available from: https://doi.org/10.11604/pamj.2014.17.146.3369
- Ibrahim RA, Cryer TL, Lafi SQ, Basha EA, Good L, Tarazi YH. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC Vet Res. 2019;15(1):1-6. Available from: https://doi.org/10.1186/s12917-019-1901-1
- Langata LM, Maingi JM, Musonye HA, Kiiru J, Nyamache AK. Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi, Kenya. BMC Res Notes. 2019;12(1):1-6. Available from: https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-019-4068-8
- Barns JN, Ezeamagu CO, Nkemjika ME, Akindele TS. Prevalence of integrons in Enterobacteriaceae obtained from clinical samples. J Microbiol Antimicrob. 2021;13(1):1-10. Available from: http://dx.doi.org/10.5897/JMA2020.433
- Nsofor CM, Tattfeng MY, Nsofor CA. High prevalence of qnrA and qnrB genes among fluoroquinolone-resistant Escherichia coli isolates from a tertiary hospital in Southern Nigeria. Bull Natl Res Cent. 2021;45:26. Available from: https://bnrc.springeropen.com/articles/10.1186/s42269-020-00475-w
- Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal and environmental ecologies. Front Microbiol. 2012;3:24. Available from: https://doi.org/10.3389/fmicb.2012.00024
- Andres P, Lucero C, Soler-Bistue A, Guerriero L, Albornoz E, Tran T, Zorreguieta A, et al. Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob Agents Chemother. 2013;57(6):2467-2475. Available from: https://doi.org/10.1128%2FAAC.01615-12
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6(10):629-640. Available from: https://doi.org/10.1016/s1473-3099(06)70599-0
- Tsekouras N, Athanasakopoulou Z, Diezel C, Kostoulas P, Braun SD, Sofia M, et al. Cross-sectional survey of antibiotic resistance in extended spectrum β-lactamase-producing Enterobacteriaceae isolated from pigs in Greece. Animals. 2022;12:1560. Available from: https://doi.org/10.3390/ani12121560
- Chah KF, Ugwu IC, Okpala A. Detection and molecular characterization of extended spectrum β-lactamase-producing enteric bacteria from pigs and chickens in Nsukka, Nigeria. J Glob Antimicrob Resist. 2018;15:36-40. Available from: https://doi.org/10.1016/j.jgar.2018.06.002
- Duru C, Nwanegbo E, Adikwu M, Ejikeugwu C, Esimone C. Extended-spectrum beta-lactamase producing Escherichia coli strains of poultry origin in Owerri, Nigeria. World J Med Sci. 2013;8(4):349-354.
- Abubakar MB, Salihu MD, Aliyu RM, Bello A, Tukur H, Shuaibu AB. Occurrence and antimicrobial resistance of ESBL-producing Escherichia coli in indigenous chickens and retailed table eggs in Sokoto Metropolis, Nigeria. Scholarly J Biol Sci. 2016;5(2):56-60. Available from: https://scholarly-journals.org/wp-content/uploads/2024/04/Abubakar-et-al.pdf